I received my Full Sequence MtDNA results in December. Of course I had no matches at this level, because so few people have tested at this level. I saw a figure of about 18,000 testers at the full sequence level (this number is probably outdated by one year or so). I would need a large number of Nicaraguans testing to find a close match at his level.
I've done a lot of research on the L2a1, and specifically the F subclade. L Haplo is African and about 55,000 years old. I found out that subclade F is anywhere from 2,000 to 10,000 years old. As you can imagine this subclade could travel far and wide throughout Africa over thousands of years. It is widespread in Northern and Central Africa, and made in roads into the Middle East. It is found in Saudi Arabia, and seems to have been spread into Europe by the Jews.
| Example 1 |
My results mismatched the L2a1f subclade by two mutations, and I have a number of extra mutations. It looks like this will lead to another subclade, which may eventually lead to a more specific location of origin for my ancestors.
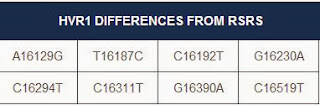 |
| Example 2 |
I found another site besides Mitosearch and Sorenson to upload my full sequence results and compare; namely the MtDNA Community.org. I found one match there. Not an exact match so we probably share an ancestors hundreds or thousands of years ago.
My best hope for a breakthrough with MtDNA would be a perfect match. I also hope we had a mutation in my line fairly recently, because that would mean we share an ancestor in the genealogical time frame.





